TARGETED IN SITU SEQUENCING ON A SUBCELLULAR LEVEL
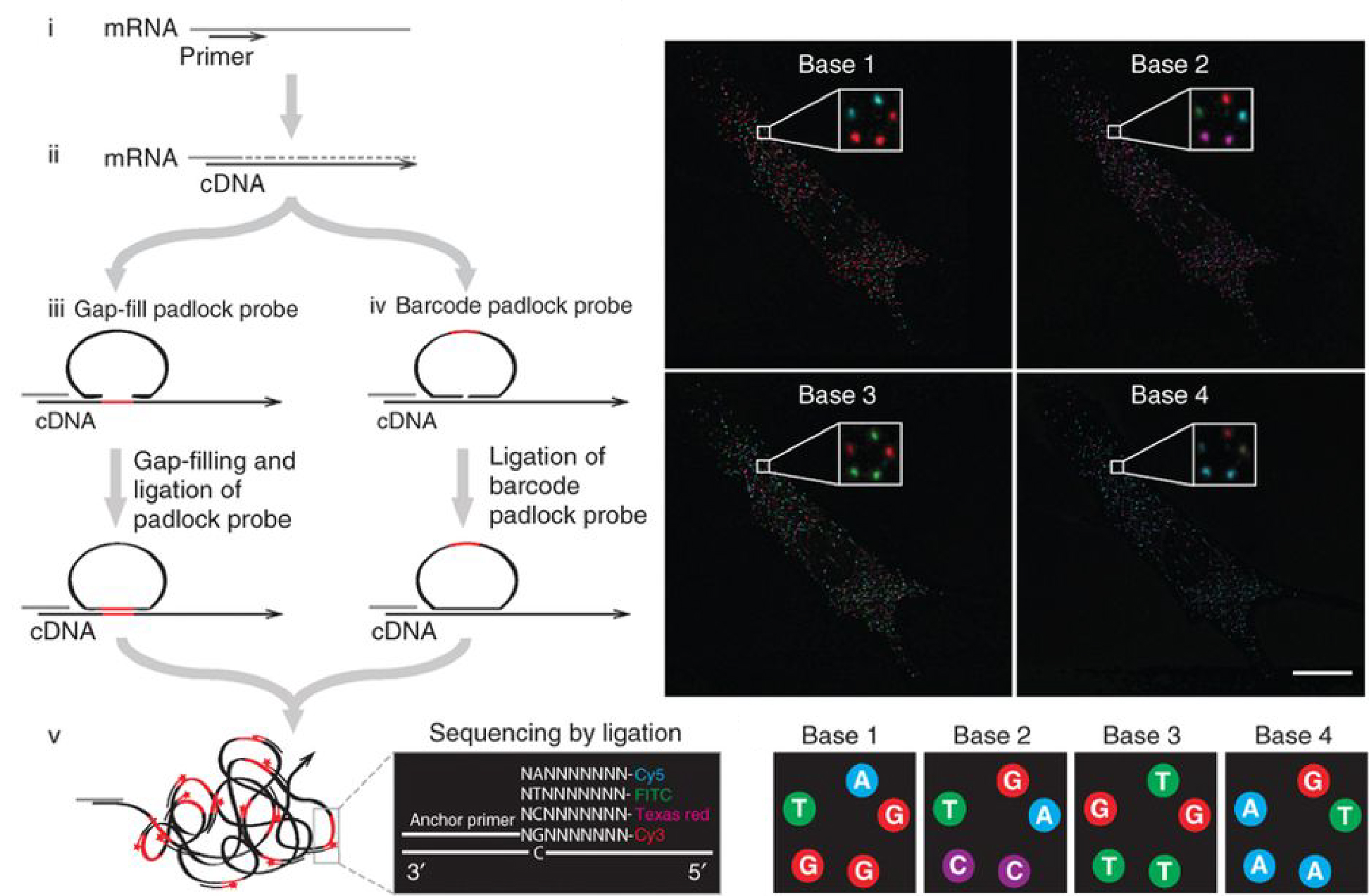
In situ sequencing is a method for targeted analysis of RNA molecules in morphologically preserved cells and tissue sections. It is based on advanced molecular tools employing nucleic acid processing enzymes and DNA circularization reactions, using padlock probes. The padlock technology enables highly multiplexed targeted ultra-deep next generation sequencing. Padlock probes combined with rolling circle amplification (RCA) provides interesting analytical advantages. First, due to the single-molecule sensitivity of these assays, they can be used for highly precise digital quantification. Second, they can be used to elicit novel magnetic or electric biosensor readouts, that can be used for hand held devices. Finally, they can be implemented in situ to detect and digitally quantify DNA and RNA sequences resolving single-nucleotide variants at micro-meter resolution, which we utilize in our in situ sequencing approach, applying next generation sequencing in situ. With this technique we can sequence DNA and RNA molecules in the preserved context of fixed cells and tissue sections and it can be applied for massively multiplexed expression profiling, splice variant mapping, and mutation detection in situ.